Y-STR profiling (accessible)
Updated 25 August 2023
FSR-GUI-0013
Issue 1
This document is issued by the Forensic Science Regulator in line with Section 9(1) of the Forensic Science Regulator Act 2021.
© Crown Copyright 2023
The text in this document (excluding the Forensic Science Regulator’s logo and material quoted from other sources) may be reproduced free of charge in any format or medium providing it is reproduced accurately and not used in a misleading context. The material must be acknowledged as Crown Copyright and its title specified.
This document is not subject to the Open Government Licence.
1. Introduction
1.1 Background
1.1.1 Humans have 23 pairs of chromosomes, 22 of which are autosomes passed on with equal likelihood to both sons and daughters, the 23rd pair being the sex chromosomes. A child inheriting an X from their father will be genetically female (denoted 46, XX) while one inheriting a Y will be male (46, XY) as a consequence of the male-determining gene (SRY) located near the end of the short arm of the Y chromosome. (Although the SRY gene is usually on the Y chromosome, it occasionally gets transferred to the X, leading to 46,XX males, whilst inactivation of SRY by mutation leads to 46,XY females (Swyer Syndrome); however, both are rare occurrences (approximately 1 in 20,000 individuals)).
1.1.2 Most genes on the Y are unique to the chromosome and are associated with fertility and the production of sperm, whilst some share homology with the X chromosome; for example, the AMEL gene (Amelogenin) used in the DNA17 test to attribute chromosomal sex. Y-STRs have similar structures and levels of variability to autosomal STRs and share many of the profile characteristics such as broadly similar levels of stutter. However, they normally display only a single allele peak as males have only one Y chromosome copy. Approximately 1 in 1,000 males have 2 identical copies of their Y chromosome (47,XYY); this trait is not inherited.
1.1.3 Unlike autosomal STRs, which are inherited independently of each other due to the random assortment of different chromosomes and recombination between paired chromosomes, the STRs on the Y chromosome are inherited from father to son as a single non-recombining set. This lack of independent inheritance means that the product rule cannot be applied to Y-STR allele frequencies. In addition, all patrilineally related males will have exactly the same combination of Y-STR alleles (known as a haplotype) unless a mutation occurs. The most common form of mutation is the alteration of the length of an STR (usually by the loss or gain of a single repeat unit).
1.1.4 Null alleles can arise in Y-STR profiles due to alteration or loss of the primer binding site, or loss of the entire Y-STR through deletion. Deletion and duplication events occur relatively frequently on the Y chromosome [1], promoted by rearrangements between the repeated structures that flank blocks of Y-STRs. As a consequence it is not uncommon for sets of adjacent STRs to be simultaneously lost or duplicated. Duplication events may only be detectable from a doubling of peak height of the duplicated STRs but eventually mutation will affect one copy and result in the appearance of a new allele, typically either one repeat larger or smaller than the original copy. Some deletions are also observed, for example, the Amelogenin Y (AMELY) deletions detected during DNA17 profiling are usually accompanied by the loss of several adjacent Y- STRs lying between the loci DYS456 and DYS19 (Figure 1); these are seen relatively commonly in males from the Indian subcontinent, at a frequency of approximately 0.02 [2].
1.1.5 Y-STR profiling has become well established in UK forensic casework since the introduction of a 12 Y-STR test (PowerPlex® Y System; Promega) in 2003, followed by a 17 Y-STR multiplex (AmpFLSTR™ Yfiler™; ThermoFisher) and subsequently by the 23 Y-STR PowerPlex® Y23 System (PPY23; Promega). Each new test added STRs while retaining those present in the preceding multiplex, the exception being Yfiler™ Plus (ThermoFisher), which includes 27 Y-STRs but omits two that are present in PPY23. Early tests were restricted to the few Y-STRs that had been characterised at the time, and included some STRs with low mutation rates (less than approximately 0.001 per STR per generation) and consequent poor discrimination power. More recent multiplexes have incorporated rapidly mutating (RM) Y-STRs, which have mutation rates greater than 0.01 per generation [3]. With the inclusion of several of these in the PPY23 and Yfiler™ Plus kits it is unusual to observe matching profiles in males not known to be closely related. Figure 1 shows the physical order of the Y- STRs, and the multiplexes that detect them. Note that locus DYS385 (present in all the multiplexes) and the multi-copy locus DYF387S1 (included in Yfiler™ Plus only) are duplicated in the vast majority of males, displaying either two separate allele peaks or a single peak of approximately double the normal height.
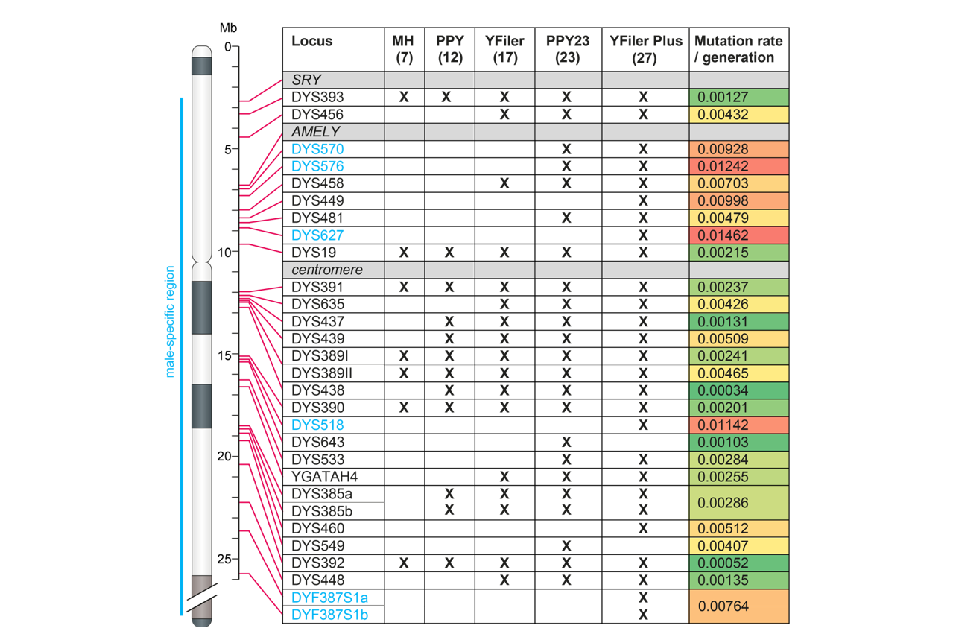
Figure 1: Physical positions of Y-STRs on the Y chromosome, inclusion in multiplexes, and mutation rates. Heat map colours show increasing mutation rates from green (low) to red (high). MH: minimal haplotype, PPY: PowerPlex Y. Physical positions of STRs are taken from Hanson and Ballantyne (2006) [4] and mutation rates are from the Y-STR Haplotype Reference Database (YHRD) Release 68; loci given in blue font (DYS570, 576, 627, 518, 387S1a and 387S1b) are considered RM Y- STRs. The positions of the SRY and AMELY genes are also shown, as is that of the centromere.
1.2 Y-STR profile considerations
1.2.1 When the Y-STR profile from a crime stain does not match that from a suspect, this excludes the suspect. However, due to the Y chromosome’s mode of inheritance, Y-STR profiling cannot distinguish between males who are paternally related unless an individual male displays a newly arisen mutation absent from his male-line relatives who would otherwise share the same profile. Even then there is a possibility that the same mutation may also have occurred independently in a more distant patrilineal relative, generating the same profile.
1.2.2 The complete absence of Y-STRs in females allows male-specific profiles to be clearly detected in female-male mixtures even when the female DNA component is very much more abundant, for example, greater than 100,000-fold excess [5]. Consequently, Y-STR profiling is invaluable in sexual assault casework where the male autosomal component cannot be isolated by alternative means such as preferential lysis [6], for example in azoospermic or vasectomised males, non-ejaculation or digital penetration. Y-STR profiles may also be detectable for hours or days after intercourse, as DNA released from degraded sperm and epithelial cells could still be present. In such cases the Y- STR profile can be compared with a reference Y-STR profile from a suspect or other persons of interest.
1.2.3 The UK currently lacks a database of either Y-STR crime stains or reference sample profiles from arrestees, and all comparisons are therefore made on a case-by-case basis. Unlike other countries (such as, Austria, China, Italy and Singapore) who have incorporated Y-STRs into their national databases, increasing the investigatory power of their DNA analysis.
1.2.4 Y-STR profiling tends to be used in challenging situations where recovery of male-related DNA components is low and the DNA generally may be degraded, therefore, it is not uncommon that the obtained profile is partial in nature. While the expectation of only a single allele at most Y-STR loci simplifies the interpretation of Y-STR profiles affected by drop-out (at least in single-source profiles, where, unlike for autosomal STRs, there is no ambiguity as to whether a single peak represents a homozygote or a heterozygote where the partner allele has dropped), the much lower discriminating power of Y-STR profiles significantly increases the likelihood that partial profiles will result in adventitious matches with unrelated males.
1.2.5 The unexpected occurrence of more than one allele at a Y-STR locus may indicate either a duplication event in the genome of the source individual, or DNA from multiple contributors. A check of whether the duplicated STRs are adjacent on the Y chromosome (Figure 1) can help to determine which explanation is more likely. The absence of a Y-STR allele in an otherwise good quality Y-STR profile indicates either a primer-binding site mutation or a deletion. When more than one Y-STR is absent, this is likely due to the same deletion event, and checking of the relative positions of the STRs on the Y chromosome can again help to determine this. However, it remains possible that alternative arrangements of the Y chromosome exist in some populations in which the relative positions of STRs as shown in Figure 1 do not apply.
1.2.6 The mode of inheritance of a Y-STR haplotype (profile) provides a useful means of establishing kinship through the male line. An identical profile is usually indicative of relatively recent shared ancestry and the more discriminating the multiplex (a combination of the number and mutation rates of the included STRs) the more recent that shared ancestry is likely to be. For example, the mean mutation rate of a haplotype defined by the PPY23 multiplex is 7.9% per generation (the sum of the per-STR mutation rates in Figure 1). Therefore two men who share a common male-line ancestor six generations ago (total, 12 generations) are extremely likely to carry different PPY23 profiles. In practice, most men observed to share a given male’s PPY23 profile will be his close male-line relatives, such as a brother, uncle, nephew, or cousin. Y-STR profiling is therefore a useful tool for establishing the significance of autosomal profile similarity in familial screens where the relationship between the individual on the database and the source of the crime stain is patrilineal.
1.2.7 Because surnames, like Y chromosomes, are passed from father to son in most societies [7], men sharing uncommon surnames are more likely to share similar Y-STR profiles than men with different surnames. The correlation between Y- STR profiles and surname is much lower for common surnames. This relationship has been used in conjunction with genetic genealogy websites to identify very distant male-line relatives. On a longer timescale, men with ancestry tracing back to the same geographic region are more likely to share similar profiles, and profiles can be grouped into clusters that are more abundant in particular continents and/or countries. However, because of recent population migration and admixture, association with geographic regions or ethnic groups is not wholly accurate. Nonetheless, the relative similarity of male profiles within such groups is significant and necessitates the use of a profile frequency reference database that reflects the likely origin of the person of interest.
2. Scope
2.1.1 The purpose of this document is to provide guidance for Y-STR analysis delivered into the criminal justice system.
2.1.2 It applies to profile (haplotype) comparison and kinship testing and does not apply to biogeographic ancestry testing.
3. Terms and definitions
3.1.1 The terms and definitions set out in the Forensic Science Regulator’s (FSR’s) Code of Practice (the Code) [14], apply to this document. Additional terms and definitions can be found in the glossary.
4. Standards and guidance
4.1.1 National and international standards for testing and calibration in laboratories (British Standard BS EN ISO/IEC 17025 [8]; International Laboratory Accreditation Cooperation ILAC G19:08/2022 [9]) provide guidance on analytical methods. However, there is much less detail for the type of interpretation of analytical results required for Y-STR analysis than for autosomal DNA analysis.
4.1.2 Scientific and technical guidelines that are relevant to (but not mandatory for) the interpretation of Y-STR profiles have been published by the International Society for Forensic Genetics (ISFG) DNA Commission [10]; [11]; [12], and by the Scientific Working Group on DNA Analysis Methods (SWGDAM) [13].
5. Y-STR profile evaluation
5.1 Single-source Y-STR profiles
5.1.1 Y-STR profiles with no more than one component (allele) at each STR (other than the locus DYS385 and other constitutively duplicated STRs) can be normally considered as a single source.
5.1.2 If the alleles are well amplified, then the Y-STR profile can be confidently assigned as originating from a single individual (although the possibility of a mixture of male paternal-line relatives should also be considered). However, in less well amplified Y-STR profiles there may be reduced confidence in assigning the male DNA detected as being from a single source.
5.1.3 Where the male DNA in a crime sample appears to be from a single source, it can be compared directly with a reference Y-STR profile from a person of interest (POI) to determine whether they match or not.
5.1.4 If the profiles match at all loci for which a designation has been made, then the crime sample may have originated from the individual who provided the reference sample, or from a member of the same paternal lineage. This finding should be evaluated as described below.
5.1.5 If the allele designations mismatch at one or more loci (and non-concordance due to differing polymerase chain reaction chemistries can be ruled out) then the male DNA recovered from the crime sample is not from the individual who provided the reference sample and should be reported as such.
5.2 Mixed Y-STR profiles
5.2.1 Y-STR profiles with more than one component (allele) at a Y-STR locus (other than DYS385, which commonly has two components) should be considered as possible mixed profiles derived from DNA from more than one male.
5.2.2 In rare cases, genomic duplication events may result in single-source profiles presenting more than one component at one or more loci, but these are unusual and nearly always restricted to a small number of loci and the duplicated peaks are well balanced.
5.2.3 As with mixed autosomal DNA mixtures, where there is a clear major contributor to the mixed Y-STR profile obtained, this major Y-STR profile can be evaluated as if it is a single-source result. The rules for safe deconvolution should be defined based on laboratory validation data. This approach is only permissible if pursued with due regard for logic, taking into account all loci, and only where it is not based on the results of the comparison of the trace with that of the POI.
5.2.4 In some cases it may be possible to condition a mixed Y-STR profile on the assumed contribution of a known male other than the POI. Commonly, this may be a known sexual partner of a female victim of a sexual offence. In such cases it may be possible to determine some or all of the Y-STR components in the mixture that originated from a male other than the known partner.
5.2.5 In both these circumstances the deduced profile can be compared directly with a reference Y-STR profile from a POI to determine whether they match or not, as described for single-source profiles.
5.2.6 However, mixed Y-STR profiles are often encountered where a clear major contributor cannot be identified. In such cases, a comparison of the mixed profile with the profile of a POI may be possible, but the findings cannot be evaluated using the statistical tools described below.
6. Y-STR statistical evaluation
6.1 Criteria for suitability for statistical evaluation
6.1.1 Only profiles interpreted as being from a single individual can currently be considered for statistical evaluation. Statistical evaluation is possible for complete or partial profiles meeting one of the following criteria. Profiles meeting none of the following criteria cannot be considered for statistical evaluation.
a. Single-source full profile.
b. Single-source partial profile.
c. Unambiguous major or minor contributor to a mixed profile. Full or partial designation of the major or minor contributor profile. Deduced profiles derived from mixed Y-STR profiles may be incomplete because of locus drop-out; where there is ambiguity in deconvolution, profiles should not be interpreted as if they were a single source.
d. Contributor to a mixture, conditioned on a known contributor. Full or partial designation of the deduced contributor profile.
6.2 Haplotype (profile) frequency databases
6.2.1 To estimate the weight of evidence in Y-STR cases where a suspect profile matches a crime-scene profile, an estimate of profile frequency needs to be determined. The requirement for large population samples to provide reasonable estimates of such frequencies means that it is common practice to use online databases to make such estimates. The most widely used example is the Y-STR Haplotype Reference Database (YHRD) [15].
6.2.2 The YHRD [16] is recommended as the default database to be used to estimate the weight of evidence for cases involving Y-STRs.
6.3 Choice of Reference Population
6.3.1 By default, the YHRD returns the number of matches in its total dataset (a worldwide population sample) for the chosen multiplex. However, it is also possible to select a ‘metapopulation’ dataset (for example, Western European metapopulation) or a national population (for example, the UK). Note, however, that the UK population here is not a subset of the Western European metapopulation, because the UK population dataset also includes Black African and South Asian individuals resident and sampled in the UK, while the metapopulation refers to the bio-geographical origin (ancestry) of the population. There is also over-representation of Black, Asian, and minority ethnic groups within the UK population dataset as a result of attempts to provide similarly sized datasets for each ethnic group.
6.3.2 Due to the general homogeneity of Y-chromosomal lineages within western Europe [17] it is appropriate to use the Western Europe metapopulation to increase the database size for comparisons with White British individuals from the UK.
6.3.3 Two alternative approaches can be used when considering which population group it is appropriate to use. The first may be referred to as “suspect anchored” whereby the population database corresponding to the ethnic group of the person of interest (POI) suspect is the default choice. The second may be referred to as “scene anchored” whereby the population database most relevant to the pool of potential perpetrators is used (thereby making no assumptions about the involvement of the POI).
6.3.4 In most cases, the scene anchored approach is preferred, i.e. focussing on the pool of potential perpetrators rather than the ethnicity of the POI. Under this approach, the Western European dataset should be used as the default dataset. The broad justification for using a scene anchored approach is that, since the majority of males resident within the United Kingdom are of Western European origin, the a priori supposed alternative source of the male DNA would most likely be from the Western European group.
6.3.5 In some situations, it may be appropriate to also consider the ‘suspect anchored’ approach, and to report a figure from the YHRD dataset thought to correspond most closely with the suspect’s ethnic appearance, in addition to the Western European figure. The decision as to whether this is appropriate or necessary will in part depend on the relative size of the additional metapopulation size to be considered and individual specifics about a case, such as whether the victim has made a physical description regarding the suspect’s ethnicity, whether the case involved a person known to the victim and the defence’ position in terms of version of events.
6.4 Choice of Method for Probability Estimation
6.4.1 The primary result returned by the YHRD is the observed number of haplotypes (profiles) matching the queried haplotype. However, this count needs to be expressed as an estimated probability of observing the haplotype in the relevant population and there are a number of possible methods to do this.
6.4.2 The YHRD website currently offers three alternative estimates based on three different published methods.
a. The n+1/N+1 pseudocounting or augmented count method (where N is the number of individual haplotypes in the relevant population dataset, and n is the number of those haplotypes matching that of the POI). This is equivalent to the frequency obtained when adding the haplotype in question to the dataset [15].
b. The ‘kappa’ correction [18] based on the observed number of singleton profiles in the database (and therefore applicable only to haplotypes not previously observed).
c. The ‘discrete Laplace’ method [19].
6.4.3 A fourth possible alternative uses a modified version of the pseudocounting method, using n+2/N+2 proposed by Balding [20].
6.4.4 Further to these different methods, alternative approaches that model haplotype distributions in living populations [21] offer a radically different solution. As these models develop, there may be a case for their adoption in casework as acceptance by the international forensic community grows.
6.4.5 It is recommended to use the n+1/N+1 approach until alternative methods emerge that have been demonstrated (through validation) to be a robust suitable replacement. This approach is in line with recommendations from the Commission of the International Society of Forensic Genetics (ISFG) (2020) [12] which states that “an alternative, easily defendable but highly conservative method is the augmented counting approach optionally with confidence interval(s) or kappa inflation. The counting approach is recommended if Y-STR profiles are partial due to degradation or include non-integer alleles”. As partial Y23 profiles are frequently encountered in forensic casework this method is preferable as a default approach to the Discrete Laplace method also recommended by ISFG but which is unsuitable for use for partial profiles. This approach is also one of the two recommended approaches for determining haplotype frequencies listed in the SWGDAM Interpretation Guidelines for Y- chromosome Y-STR typing 2022 [13].
6.5 Choice of multiplex for evaluation purposes
6.5.1 The YHRD database offers a number of different search configurations whereby different data sets of YSTR profiles and different search profile configurations can be selected. This is in order to maximise the information returned from the complex total dataset which made up of profiles generated from several different, though overlapping (in terms of loci) multiplexes.
6.5.2 Release 67 of YHRD (Feb 2022) updated and improved the search options available on YHRD and these recommendations are based on those search configurations (i.e. release 67 or later).
6.5.3 There are two useful and valid options for YHRD searches using a full or partial Y23 profile.
6.5.4 The first is to limit the dataset to be searched to the Y23 dataset only. This search will only include reference samples for which a full Y23 profile is present on YHRD. This dataset is necessarily smaller than the complete YHRD dataset as it does not include the profiles derived from the Yfiler or Yfiler plus kits (which do not include all of the Y23 loci).
6.5.5 The second search type is a modified search to enable a larger YHRD dataset to be searched.
6.5.6 UK forensic units primarily use a 23-locus Y-STR system (PPY23), however it is possible to modify a YHRD search to include in the search the larger set of 17- locus Y17 profiles. This Y17 dataset includes all profiles which include the Y17 loci (so Yfiler + Powerplex Y23 + Yfiler plus profiles). The “transient” search option available in YHRD requires the user to select the PowerPlex Y23 kit option, but the Y17 dataset option to maximise the information returned from the search. This search configuration returns all profiles from the large Y17 dataset which match against the Y23 search profile.
6.5.7 Choosing to search databases with different groups of loci is referred to specifically in the Scientific Working Group on DNA Analysis Methods guidelines [13], which support this approach, stating: “Due to the challenge of small database sizes for the larger multiplex systems, it is acceptable to perform additional searches of the population database using reduced locus sets in an attempt to obtain the most informative result for that combination of evidence and population database profiles”.
6.5.8 Given the inverse relationship between profile information content (number of loci) and number of records, there is no clear-cut position on which profile datasets are most appropriate. The following recommendation takes account of this by devolving this decision to a case-by-case assessment.
a. Profile probabilities should be calculated using both the Y23 search profile and Y23 data set search; and the Y23 search profile and Y17 data set search.
b. The lowest frequency (i.e. the most discriminating) should be reported.
7. Reporting Y-STR results
7.1 Statistical evaluations
7.1.1 When assessing the probability of observing a Y-STR profile in a relevant population using the YHRD, the following information should be included within reports and statements:
a. That the Y-STR profile obtained from the evidential sample matches the person of interest (POI).
b. That any male belonging to the same paternal lineage as the POI will also be likely to match
c. That the YHRD was used to estimate the frequency of the Y-STR profile, citing in addition,
- i. the (meta)population used
- ii. that if the profile was re-searched at a later date the probability reported might change.
- iii. The number of matching profiles seen in the population searched, and the size of that population dataset, the date of the search, and the YHRD release number may also be included.
d. The results may be presented as a either a likelihood ratio or as a relative frequency.
e. An activity level interpretation, if at all possible.
7.1.2 When expressing a sub-source level conclusion as a likelihood ratio (LR), the alternative propositions being considered must first be set out.
7.1.3 An example of the possible wording is shown below: * Proposition 1: The source of the male DNA is [Suspect name] (or a close paternal-line male relative of his). * Proposition 2: The source of the male DNA is an unrelated male from the [reference population].
7.1.4 An example of possible wording when providing the LR in a report or statement is shown below:
It is estimated that the male DNA from [the crime stain sample] is approximately LR times more likely if the first proposition were true rather than if the second proposition were true.
7.1.5 If it is the policy of the forensic unit to convert numeric findings using the standard verbal scale of support, then the relevant point on that scale may also be reported in addition to the numerical finding.
7.1.6 An example of the wording that could be used (for sub-source level) is shown below:
In my opinion, the scientific findings provide [degree of support] for the view that [Suspect name] (or a close paternal-line male relative of his) deposited the male DNA on the [crime stain item] rather than an unknown unrelated man.
7.2 Combining Y-STR and autosomal STR statistics
7.2.1 In some cases, it may be desirable to combine the statistical evaluations of Y- STR and autosomal STR results from the same case to provide a combined likelihood ratio. This is supportable if there is a reasonable expectation of genetic independence of the two marker sets. Such independence studies and associated considerations of this approach have been reported by Walsh et al. [22] and by Buckleton and Myers [23] who report only mild effects resulting from the assumption of independence.
7.3 Alternatives to statistical evaluation
7.3.1 In cases where a statistical evaluation is not possible (e.g. for many mixed Y- STR profiles), a comparison of the reference Y-STR profile of a POI to the crime profile may, in some cases, still have been carried out. In doing so, the scientist may reach a conclusion that the POI cannot be excluded as a possible contributor to the Y-STR profile obtained from the crime stain sample. In this situation, there are a number of possible approaches to reporting this outcome.
a. If no statistical evaluation is possible, the forensic unit may report the profile as unsuitable for further evaluation and make no comment about the possibility of contribution, nor state the name of the POI.
b. The forensic unit may only provide an expression of the possibility that the POI contributed to the mixture if it is presented in a manner that does not favour the prosecution; such an expression is likely to be uninformative. If an assessment of evidential weight is not possible, the scientist should make it clear that they can give no guidance to the court with regard to probative value.
c. The forensic unit may provide a qualitative or subjective evaluation, if it is supported by scientific experimentation, such as non-contributor testing (whereby a large number of random profiles from individuals not associated with the investigation are compared to the evidential mixed profile to determine if they would have been considered as ‘possible’ contributors themselves). If such analysis is conducted it may be reasonable to conclude that the findings in the case are more likely if the POI (or a close paternal-line male relative of his) had contributed DNA to the mixed Y-STR profile obtained rather than if someone selected at random from the wider general population had contributed DNA to the mixed Y-STR profile. When providing a qualitative or subjective evaluation, the scientist may or may not be able to provide a level of support for their finding. This will depend on the specific qualities of the Y- STR profile obtained from the crime stain sample.
d. Andersen and Balding (2017) [21] demonstrate that the chance of a randomly selected man matching a POI is negligible. Andersen and Balding (2019) [22] show that a two-male mixture that includes the profile of a POI has almost exactly the same evidential value as a single contributor match to the POI.
7.4 Future Challenges
7.4.1 The statistical evaluation of Y-STR profiles differs significantly from that of autosomal profiles. In order to report and evaluate Y-STR results in a robust manner and to achieve the maximum evidential value from such results, a number of requirements can be identified.
a. Provision of a larger reference population of UK-resident populations, preferably collected across different regions of the UK and with their geographical provenance recorded.
b. Further investigation and guidance from the international forensic community on appropriate methods to estimate profile probabilities and report weight of evidence for single-source profiles.
c. Further investigation and guidance from the international forensic community on appropriate methods to statistically evaluate mixed profiles where a clear single-source contributor cannot be deduced. This may include the development of probabilistic models applicable to Y-STR profiles.
8. Y-STR for kinship investigations
8.1 Background
8.1.1 The pattern of patrilineal inheritance of Y-STR haplotypes can assist in kinship investigations. Simplistically, members of the same paternal lineage are (barring mutation) expected to share a common Y-STR haplotype. Therefore the observation of a shared haplotype supports the hypothesis that two men are from the same paternal lineage rather than that they are unrelated.
8.1.2 Although this simple assumption of matching haplotypes between male relatives is largely upheld, the occurrence of mutations of one or more Y-STR loci during meiosis in the spermatogenesis process may result in one or more differences in allele designations between male relatives. The more meioses that separate the two individuals, the greater the chance that at least one of the Y-STR loci will undergo a germline mutation.
8.1.3 The probability of two male relatives having different haplotypes at one or more loci is dependent on
a. the number of STR loci being tested,
b. the mutation rates for those STR loci, and
c. the degree of relatedness of the two men (expressed in terms of the number of meiotic events separating the two men).
8.1.4 Thus, a father and son (with a single meiotic transmission separating them) will be more likely to have identical haplotypes than two full brothers (who are separated by two meioses) or two first cousins (who are separated by 4 meioses).
8.1.5 Mutation rates for the relevant PPY23 Y-STR loci have been determined and published, and it is therefore possible to compute the probability that pairs of related men with differing degrees of relatedness will possess matching haplotypes, or will differ at one, two or more (n) loci.
8.2 Statistical evaluation in kinship investigations
8.2.1 Alleged familial relationships (between two male A and B) may be evaluated statistically by considering a pair of alternative propositions, for example:
Hp: A and B are related as father and son (or any alternative specified relationship)
Hd: A and B are unrelated men
8.2.2 In the example above, assuming that A and B have full PPY23 haplotypes matching at all loci, then the LR numerator, p(E|Hp) needs to be computed on the basis that there was 1 meiotic transmission and that no mutations occurred. For each locus in the haplotype set, that is computed as 1 – u (where u is the applicable mutation rate for that locus).
8.2.3 The LR denominator, p(E|Hd) is the estimated frequency of the haplotype seen in the son determined from the YHRD database.
8.2.4 A Kinship Analysis tool that will perform this calculation is included in the YHRD toolkit . Please see the YHRD website [16] for the tool specifications.
9. Quality assurance for Y-STR profiling
9.1 Quality assurance checks
9.1.1 Quality assurance (QA) checks on Y-STR profiles should comply as much as possible with the requirements set out in the Code. Forensic units should take the same measures to minimise the risk of contamination in Y-STR profiling as when producing autosomal STR profiles.
9.1.2 Forensic units should take the same measures, where possible, to identify unknown Y-STR profiles in Y-STR profiling as when undertaking autosomal STR profiling. This includes the creation and maintenance of elimination databases. It is likely that only a local elimination database of Y-STR profiles can be maintained, including such profiles from male personnel, visitors to laboratory areas (for example, engineers), and unsourced contaminants. Given the risk of contamination, special consideration should be given to obtaining the Y-STR profiles of practitioners who carry out incident scene examination and/or those and forensic medical examiners working in sexual assault referral centres, or similar facilities.
9.2 Co-processing of quality assurance controls
9.2.1 Where possible, extraction negative controls should be processed alongside Y- STR samples. These will provide assurance for the Y-STR process. Negative controls can also provide information on the rate of drop-in seen within the Y- STR profiling method.
9.2.2 In some instances, the DNA extract from controls processed with samples during autosomal STR profiling may be consumed to such an extent (for example, in the investigation of a suspected contamination event) that insufficient DNA extract remains for co-processing of the controls with the samples using Y-STRs. In such instances, the reason that QA controls from autosomal DNA profiling were not reworked with Y-STRs alongside the co- processed samples must be recorded.
9.2.3 Validation of Y-STR profiling may provide assurance that, where no autosomal STR profile is obtained from a negative control using the kit used for regular processing of crime stains, then the expectation that no Y-STR profile will be obtained either is correct. Where such a demonstration has been made during validation, or derived from processing a sufficient number of controls with both autosomal and Y-STR kits, consideration may be given to not co-processing extraction negative controls with Y-STR samples.
9.2.4 Where an unsourced contaminant (sufficient for retention on a local elimination database) is identified in a QA control sample in autosomal STR profiling, and the profile is not certain to have derived from a female source, a Y-STR profile should be produced from the control where possible. This should be done even if no samples co-processed with that control have been subject to Y-STR profiling, as samples from other batches may be processed in the same laboratory or with the same consumables.
10. Investigation of potential contamination in Y-STR profiles
10.1.1 It is likely that not every sample generated from crime stains and associated reference samples will automatically undergo Y-STR profiling. A Y-STR profile may be generated from a crime stain sample and not found to match a nominal from whom a reference sample has been submitted. Considered to be of unknown origin, such a sample may originally have been processed for the purpose of generating an autosomal STR profile alongside samples for which no Y-STR profile was requested or produced. If the unknown Y-STR profile is the result of cross-contamination from another sample co-processed during autosomal STR profiling, it is possible that the contamination event will go undetected if the sample from which the contaminant derived has not also been Y-STR profiled.
10.1.2 This restricts the investigation of potential cross-contamination events leading to the generation of a contaminant Y-STR profile. Forensic units should not produce Y-STR profiles for any crime stain or reference samples other than those for which a request is submitted. Generating Y-STR profiles from samples in other cases, where no such profiling was requested, will require the disclosure of any Y-STR profiles and may lead to further Y-STR profiling of samples from nominals and other crime stains that were also not requested.
Aside from the legal complexity of Y-STR profiling that was not requested or required, such investigative Y-STR profiling would consume DNA recovered from a sample that might be required for re-work with the regular testing method.
10.1.3 In some instances, for Y-STR profiles for which the origin is unknown, comparison between the autosomal STR profile obtained for that sample with other autosomal STR profiles obtained from co-processed samples can be considered. This may be carried out with the understanding that, due to differences in the amount of template DNA input, or to the presence of large amounts of female DNA in the autosomal DNA reaction, no evidence of sample- to-sample contamination may be found by comparing autosomal STR profiles even where sample-to-sample contamination has occurred and resulted in a contaminant Y-STR profile.
10.1.4 Forensic units should investigate the origin of any unknown Y-STR profile obtained as much as possible. The limitations of the investigation should be noted along with any findings. Given the above limitations, contamination can almost never be ruled out as the source of an unknown Y-STR profile; this does not justify failure to investigate the origin of unknown Y-STR profiles.
10.2 Sharing unsourced contaminant Y-STR profiles
10.2.1 The National DNA Database provides a mechanism for retaining and sharing information on unsourced contaminant profiles generated with approved autosomal STR kits. No such mechanism is available for the sharing of Y-STR unsourced contaminant profiles. Forensic units should circulate a list of points of contact for the purposes of sharing such profiles.
10.3 Y-STR elimination databases
10.3.1 As for autosomal DNA profiling, an elimination database containing the Y-STR profiles of personnel should be created and maintained for comparison with unknown Y-STR profiles prior to their being reported or loaded to a DNA database.
10.4 Creation of a Y-STR elimination database
10.4.1 The nature of Y-STR profiles, and the likely approach taken to routinely generating such profiles in the laboratory, creates some instances where the approaches taken to using a Y-STR elimination database (YED) must differ from using an elimination database containing autosomal STRs. These issues include shared ancestry, fertility, sex-reversal syndromes and gender.
10.4.2 Y-STR profiles from personnel may reveal facts about them that should not be made known to colleagues and about which, in some cases, they themselves may be unaware.
a. Different Y-STR profiles obtained from males believing themselves to be from the same paternal line (for example, brothers) may reveal a difference in their paternity.
b. The absence of several loci from a Y-STR profile may be indicative of a deletion if the loci are adjacent to each other on the Y-chromosome. Genes linked to fertility may also have been deleted, perhaps preventing the individual from fathering children.
10.4.3 In rare sex-reversal syndromes chromosomal sex is discordant with phenotypic sex. For example, an individual with androgen insensitivity syndrome carries a Y-chromosome, but is phenotypically female. In autosomal DNA profiling such an individual will show an Amelogenin Y (AMELY) result, and would provide a full Y-STR profile if tested.
10.4.4 The forensic unit should have policies and procedures in place regarding the personnel that will be required to provide DNA profiles for elimination databases. These personnel should be informed of the purpose of providing DNA - to generate and retain the resultant DNA profiles for comparison in order to detect cross-contamination. Personnel should also be told that Y-STR profiling will be progressed where AMELY is observed in the autosomal profile, or for those presenting themselves as male, and that Y-STR profiling will reveal the chromosomal status of the donor.
10.4.5 Obtaining Y-STR elimination profiles from transgender personnel must be considered carefully. Employers should be mindful of any legal protections for such individuals under the Equality Act 2010 [23].
10.4.6 In anticipation of these scenarios, forensic units must carefully consider how to use and manage Y-STR profile information obtained from personnel and visitors for the purposes of elimination, and especially how best to restrict access to any database holding Y-STR profiles so as to avoid revealing personal information to other personnel. It is therefore likely that the YED must be more closely protected than the regular autosomal STR elimination database. Creation of a separate, local Y-STR elimination database by each forensic unit will make it easier to enact protective measures. Communicating matches without naming the individual on the YED to whom the profile matched will ensure that anonymity is maintained.
10.4.7 Forensic units may use personnel as volunteers donors with consent for validation, research, and blind testing of the processes and systems in place. The use of personnel for repeated quality assurance batch controls should be avoided. Care should be taken when requesting donor samples of any sort for producing Y-STR profiles where a potential donor has a Y-STR profile that may involve a genetic privacy issue. Repeat use of that donor will draw attention to the affected profile. Where the issue is with fertility, any impact on the quality of sperm donated for use as control or experimental material for non-DNA processes should also be considered.
10.5 Searching Y-STR elimination databases
10.5.1 Careful consideration should be given to the minimum number of alleles used to search the YED. Searching supposed contaminant profiles that are very partial might result in adventitious matches and unnecessary investigations. Searching only nearly complete profiles may prevent detection of genuine contamination. Presumptive alleles not labelled or included in the search should also be compared against any matches obtained.
10.5.2 Unlike complete autosomal STR profiles commonly generated from crime stains, many individuals in a population may share the same Y-STR profile. Also, the size of a local YED may not be very different, perhaps only an order of magnitude, from the size of any database, or subset thereof, used to determine the statistical significance of a Y-STR profile. Given the similarity between Y- STR profiles, partial profiles may produce several matches when searched on a YED. As with autosomal DNA elimination databases, the larger a YED is, the more likely a matching profile will be found. However, a match between an unknown Y-STR profile and a record on a local YED may not be as significant as a match between autosomal STR profiles.
10.5.3 As patrilineal male relatives are expected to share the same Y-STR profile, search of unknown profiles against a local YED may be more likely to produce matches where a crime has been committed in the same community in which those sampled, or their relatives, have lived. There is also some evidence for a correlation between Y-STR haplotypes and surnames for which a single origin is likely. Forensic units should understand that a local YED may operate as a ‘de facto’ mini-database, highlighting possible familial connections.
11. Guidelines for interpreting the presence and designation of peaks
11.1.1 As with autosomal short tandem repeat (STR) typing, forensic units conducting Y-STR analyses should characterise the performance of their systems and the profiles produced as part of their validation process. These data should be used to develop appropriate guidelines for the interpretation of such profiles.
11.1.2 Forensic units should consider the following thresholds and parameters.
a. Analytical threshold: The peak height below which alleles cannot safely be designated. This threshold may be dependent on the multiplex kits used, the amplification and detection systems in place, and other factors that may impact on the baseline noise and the signal strength. An appropriate threshold should be determined by each forensic unit.
b. Stutter: Polymerase chain reaction amplification of Y-STRs is likely to generate stutter artefacts similar to those seen with autosomal STRs. These will most commonly be one repeat unit shorter than the primary allele, but additional stutter products (for example, two repeats smaller or one repeat larger) may also be observed. Interpretation guidelines to help to identify stutter may be developed at the multiplex level, the locus level or at the level of individual alleles.
c. Stochastic threshold: For an autosomal locus, this is defined as the peak height below which a single allele peak at a locus cannot safely be designated as a homozygote at an autosomal locus (because of possible allele drop-out of a second allele). Most Y-STRs commonly used for forensic analyses are single-copy loci present only once in the male genome. A stochastic threshold is not applicable to single copy loci. However, for duplicated Y-STR loci (such as locus DYS385a,b) it is appropriate that a stochastic threshold is determined.
d. Peak height ratio / heterozygote balance: Thresholds based on the ratio of two allele peaks presumed to be heterozygous alleles from the same individual provide useful guidance in the interpretation of autosomal STRs. Peak height ratios are not applicable to single copy loci. However, for duplicated Y-STR loci (such as DYS385a,b) it is appropriate that the characteristics of peak height ratio of the two alleles are determined and appropriate thresholds developed.
e. Non-specific artefacts: Y-STR multiplexes designed for forensic analysis are highly specific for the intended Y-chromosome loci. However, it has been demonstrated that in the presence of very large excesses of human genomic DNA (usually female) low levels of non-specific amplification products may be observed (Moore et al., 2016) [17]. These appear in characteristic positions likely to be dependent on the exact multiplex kit used. Forensic units should have appropriate guidelines for identifying and reporting such artefacts. Forensic units should also have guidance to identify other non-allelic artefact peaks that may occur in Y-STR profiles.
f. Deletions and duplications: The Y chromosome is prone to copy number variation including deletion and duplication of stretches of the genomic sequence, which may impact on STR loci (see 1.1.3 and 1.2.6). Forensic units should have appropriate guidelines for identifying and reporting such occurrences.
12. Modification
12.1.1 This is the first issue of this document.
12.1.2 The Regulator uses an identification system for all documents. In the normal sequence of documents this identifier is of the form ‘FSR-#-###’ where (a) (the first ‘#’) indicates a letter to describe the type of document and (b) ‘###’ indicates a numerical, or alphanumerical code to identify the document. For example, this document is FSR-GUI-0013, and the ‘G’ indicates that it is a guidance document. Combined with the issue number this ensures that each document is uniquely identified.
12.1.3 If it is necessary to publish a modified version of a document (for example, a version in a different language), then the modified version will have an additional letter at the end of the unique identifier. The identifier thus becoming FSR-#-####.
12.1.4 In all cases the normal document bearing the identifier FSR-#-### is to be taken as the definitive version. In the event of any discrepancy between the normal version and a modified version then the text of the normal version shall prevail.
13. Acknowledgements
13.1.1 The guidance in this document is based on published work. There has also been widespread consultation with all of the major providers of forensic science services in the UK and Ireland and academia via the Forensic Science Regulator DNA Specialist Group. The FSR would like to thank Cellmark Forensic Services, Eurofins Forensic Services, Forensic Science Ireland, Forensic Science Northern Ireland, Key Forensic Services Ltd, King’s College London, Scottish Police Authority Forensic Services, University of Leicester.
14. Review
14.1.1 This published guidance will form part of the review cycle as determined by the Forensic Science Regulator.
14.1.2 The Forensic Science Regulator welcomes comments. Please send them to the address as set out on the Forensic Science Regulator home page, or email: [email protected].
15. References
-
M. A. Jobling, “Copy number variation on the human Y chromosome,” Cytogenet, vol. 123, no. 1-4, pp. 253-62, 2008.
-
M. A. Jobling, I. C. Lo, D. J. Turner, G. R. Bowden, A. C. Lee, Y. Xue, D. Carvalho- Silva, M. E. Hurles, S. M. Adams, Y. M. Chang, T. Kraaijenbrink, J. Henke, G. Guanti and B. v. O. R. A. M. R. J. d. K. P. a. T. McKeown, “Structural variation on the short arm of the human Y chromosome: recurrent multigene deletions emcompassing Amelogenin,” Human Molecular Genetics, vol. 16, no. 3, pp. 307-316, 2006.
-
K. N. Ballantyne, M. Goedbloed, R. Fang, O. Schaap, O. Lao, A. Wollstein, Y. Choi, K. van Duijn, M. Vermeulen, S. Brauer, R. Decorte, M. Poetsch, N. von Wurmb-Schwark, P. de Knijff, D. Labuda, H. Vézina, H. Knoblauch, R. Lessig and L. Rower, “Mutability of Y-chromosomal microsatellites: rates, characteristics, molecular bases, and forensic implications,” American Journal of Human Genetics, vol. 87, no. 3, pp. 341-353, 2010.
-
E. K. Hanson and J. Ballantyne, “Comprehensive annotated STR physical map of the human Y chromosome: Forensic implications,” Legal Medicine, vol. 8, pp. 110-120, 2006.
-
D. Moore, T. Clayton and J. Thomson, “Description of artefacts in the PowerPlex Y23((R)) system associated with excessive quantities of background female DNA,” Forensic Science International: Genetics, vol. 24, pp. 44-50, 2016.
-
P. Gill, A. Jeffreys and D. J. Werrett, “Forensic application of DNA ‘fingerprints’,” Nature, vol. 318, pp. 577-579, 1985.
-
T. King and M. Jobling, “What’s in a name? Y chromosomes, surnames and the genetic genealogy revolution,” Trends in Genetics, vol. 25, pp. 351-360, 2009.
-
British Standard, BS EN ISO/IEC 17025, General requirements for the competence of testing and calibration laboratories.
-
ILAC, “G19:06/2022 Modules in a Forensic Science Process,” 2022. [Online]. Available: https://ilac.org/publications-and-resources/ilac-guidance-series/. [Accessed 3 8 2023].
-
P. Gill, C. Brenner, B. Brinkmann, B. Budowle, A. Carracedo, M. A. Jobling, P. de Knijff, M. Kayser, M. Krawczak, W. R. Mayr, N. Morling, B. Olaisen, V. Pascali, M. Prinz, L. Roewer, P. M. Schneider, A. Sajantila and C. Tyler-Smith, “DNA Commission of the International Society of Forensic Genetics: Recommendations on forensic analysis using Y chromosome STRs,” Forensic Science International, vol. 124, pp. 5- 10, 2001.
-
L. Gusmão, J. Butler, A. Carracedo, P. Gill, M. Kayser, W. Mayr, N. Morling, M. Prinz, L. Roewer, C. Tyler-Smith and P. Schneider, “DNA Commission of the International Society of Forensic Genetics (ISFG): An update of the recommendations on the use of Y-STRs in forensic analysis,” Forensic Science International, vol. 157, pp. 187-197, 2006.
-
L. Roewer, M. Anderson, J. Ballantyne, J. Butler, A. Caliebe, D. Corach, M. D-Amato, L. Gusmão, Y. Hou, P. de Knijff, W. Parson, M. Prinz, P. Schneider, D. Taylor, M. Vennemann and S. Willuweit, “DNA Commission of the International Society of Forensic Genetics: Recommendations on forensic analysis using Y chromosome STRs (ISFG): recommendations on the interpretation of Y-STR results in forensic analysis.,” Forensic Science International: Genetics, vol. 48, p. 10230, 2020.
-
Scientific Working Group on DNA analysis methods (SWGDAM), “Interpretation guidelines for Y-chromosome STR testing,” 2022. [Accessed 3 8 2023].
-
Forensic Science Regulator, “Code of Practice,” 2023. [Accessed 3 8 2023].
-
S. Willuweit and L. Roewer, “The new Y chromosome Haplotype Reference Database,” Forensic Science International: Genetics, vol. 15, pp. 43-48, 2015.
-
S. Willuweit and L. Roewer, “Y-Chromosome haplotype reference database,” 2023. [Online]. Available: https://yhrd.org/. [Accessed 3 8 2023].
-
L. Roewer, P. Croucher, S. L. T. Willuweit, M. Kayser, R. Lessig, P. de Knijff, M. Jobling, C. Tyler-Smith and M. Krawczak, “Signature of recent historical events in the European Y-chromosomal STR haplotype distribution.,” Human Genetics, vol. 116, no. 4, pp. 279-291, 2005.
-
C. Brenner, “Fundemental problem of forensic mathematics - The evidential value of a rare haplotype.,” Forensic Science International: Genetics, vol. 4, pp. 281-291, 2010.
-
M. Anderson, P. Svante and N. Morling, “The discrete Laplace exponential family and estimation of Y-STR haplotype frequencies,” Journal of Theoretical Biology, vol. 329, pp. 39-51, 2013.
-
D. Balding, Weight-of-evidence for Forensic DNA Profiles, Chichester: John Wiley & Sons Ltd, 2005.
-
M. Anderson and D. Balding, “How convincing is a matching Y-chromosome profile?,” PLOS Genetics, vol. 13, no. 11, p. e1007028, 2017.
-
B. Walsh, A. Redd and M. Hammer, “Joint match propabilities for Y-chromosomal and autosomal markers,” Forensic Science International, vol. 174, pp. 234-238, 2008.
-
J. Buckleton and S. Myers, “Combining autosomal and Y chromosom match probabilities using coalescent theory,” FSI Genetics, vol. 11, pp. 52-55, 2014.
-
M. Anderson and D. Balding, “Y-profile evidence: Close paternal relatives and mixtures,” Forensic Science International: Genetics, vol. 38, pp. 48-53, 2019.
-
Equality Act, The Stationary Office, 2010.
-
Moore, D., Clayton, T,, Thomson, J., “Description of artefacts in the PowerPlex Y23((R)) system associated with excessive quantities of background female DNA,” Forensic Science International: Genetics 2016; 24: 44-50., vol. 24:, pp. 44-50, 2016.
16. Abbreviations and acronyms
AMELY: Amelogenin Y
BS EN: British Standard European Norm
DNA: Deoxyribonucleic acid
DNA17: 17 STR loci system including the gender marker Amelogenin
FSR: Forensic Science Regulator
IEC: International Electrotechnical Commission
ILAC: International Laboratory Accreditation Cooperation
ISFG: International Society for Forensic Genetics
ISO: International Organization for Standardization
MH: Minimal Haplotype (9 Y-STR loci)
POI: Person of interest
PPY23: PowerPlex® Y23 System (23 Y-STR loci)
QA: Quality assurance
STR: Short tandem repeat
SWGDAM: Scientific Working Group on DNA Analysis Methods
YED: Y-STR elimination database
YHRD: Y-STR Haplotype Reference Database
17. Glossary
Amelogenin
A set of proteins encoded by a single-copy gene located on the X chromosome (AMELX) and on the male-specific region of the Y chromosome (AMELY). A 6 base pair deletion in AMELX enables each to be visualised after electrophoretic separation, informing the determination of sex.
Artefact
A ‘nuisance’ peak in a profile; often associated with the amplification and detection processes, such as a spike, dye blob, or spectral pull-up. Artefacts do not represent genuine alleles and are screened out by the scientist or the software.
Autosomal DNA
DNA from the 22 pairs of non-sex chromosomes.
Duplication Events
Genetic duplication of stretches of the genomic sequence. Most duplication events result in two alleles at a single-copy locus that differ by a single repeat unit.
These may result in single source profiles having more than one component at one or more loci.
Locus (plural loci)
The specific genetic location of an allele on a chromosome. Short tandem repeats are examples of loci that are used in forensic science because they are polymorphic and are therefore highly discriminatory when several are analysed in combination to generate a DNA profile.
Mixture
A DNA profile that contains more designated alleles than would be expected if there were only one contributor to the sample.
Negative control (Blank)
Contains the analyte at a concentration below a specified limit. The intention is that no DNA is present and no profile or alleles above the drop-in rate are expected.
Partial profile
A DNA profile that is missing one or more alleles from the donor. This can be because the DNA has been degraded, or because DNA is present at such low levels that accurate marker information cannot be obtained.
Patrilineal
Relating to descent through the male line. Grandfather, father and son share a patrilineal relationship.
Stutter
An artefact of the amplification process that leads to smaller peaks close to the main allelic peak. The most common stutter peak is one repeat unit smaller than the allelic peak (for a tetranucleotide short tandem repeat, - 4). Stutters with other numbers of repeats are also possible, but less common. Over-stutters are one repeat unit larger than the allelic peak (+4).
Y-STR profile
A short tandem repeat (STR) profile derived from the combination of several STRs on the Y-chromosome, also known as a Y-STR haplotype.
18. Further reading
Peach, C. (2006) ‘South Asian migration and settlement in Great Britain, 1951–2001’, Contemporary South Asia, 15:2, pp 133-146, DOI: 10.1080/09584930600955234.
The Royal Society and the Royal Society of Edinburgh (2017) Forensic DNA Analysis – A primer for courts. [Accessed 10 03 2021].
UKAS® (2016) UKAS Policy on Participation in Proficiency Testing, TPS 47, edition 3, issued November 2016. United Kingdom Accreditation Service [Accessed 10 03 2021].

